Visualization of data and simulations
In this notebook, we illustrate the visualization functions of petab.
from petab.visualize import plot_with_vis_spec, plot_without_vis_spec
folder = "example_Isensee/"
data_file_path = folder + "Isensee_measurementData.tsv"
condition_file_path = folder + "Isensee_experimentalCondition.tsv"
visualization_file_path = folder + "Isensee_visualizationSpecification.tsv"
simulation_file_path = folder + "Isensee_simulationData.tsv"
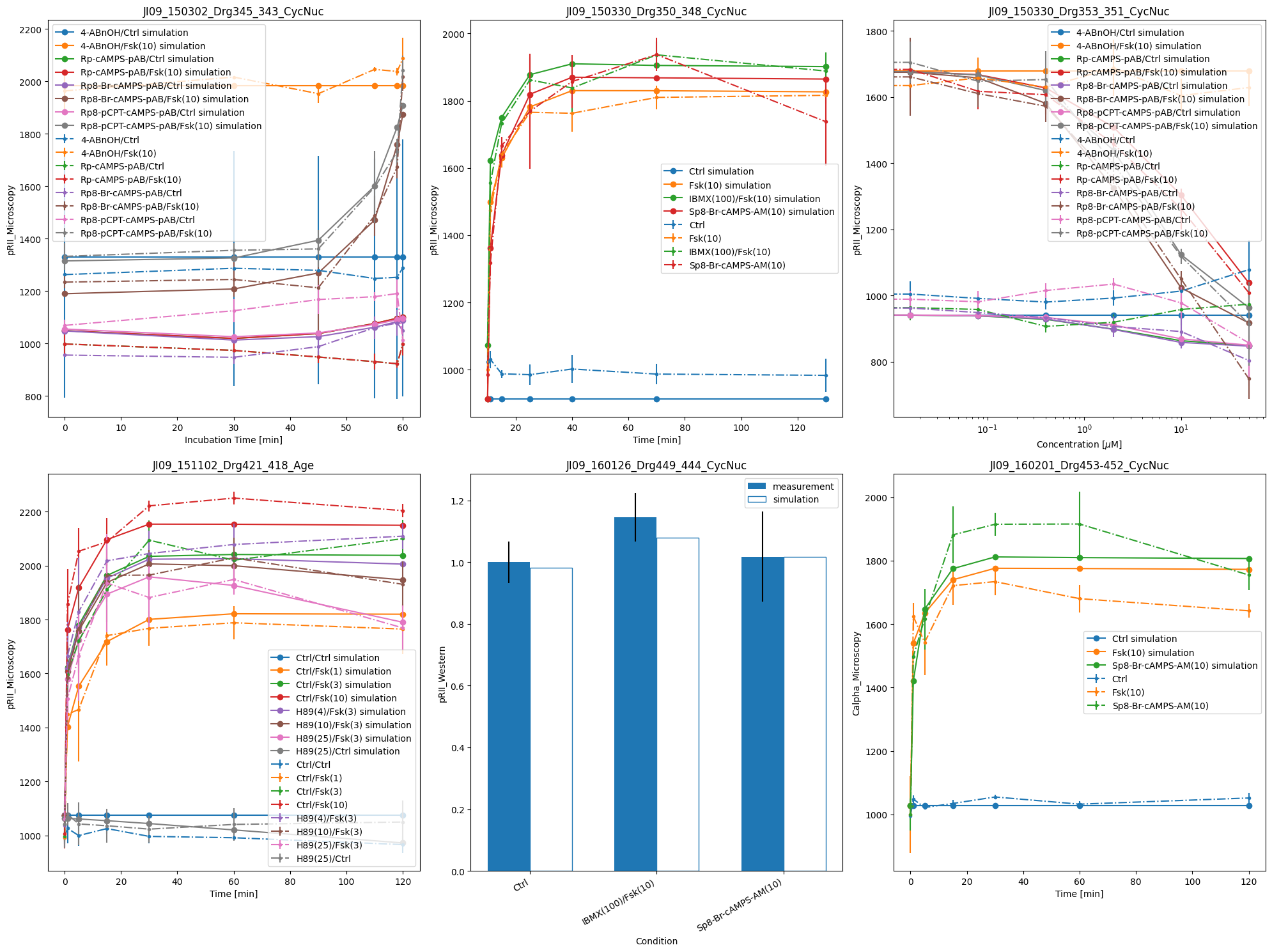
ax = plot_with_vis_spec(
visualization_file_path,
condition_file_path,
data_file_path,
simulation_file_path,
)
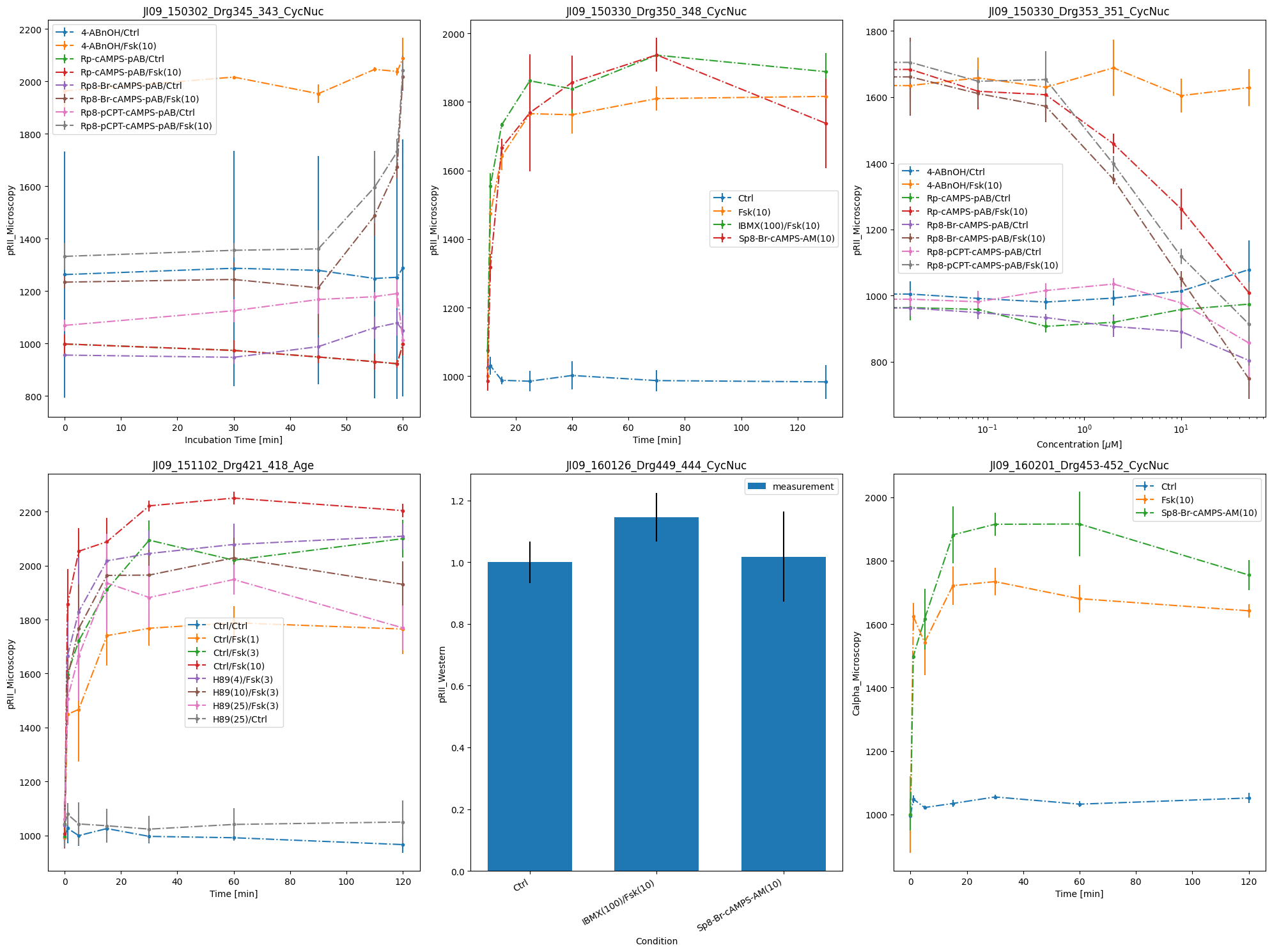
Now, we want to call the plotting routines without using the simulated data, only the visualization specification file.
ax_without_sim = plot_with_vis_spec(
visualization_file_path, condition_file_path, data_file_path
)
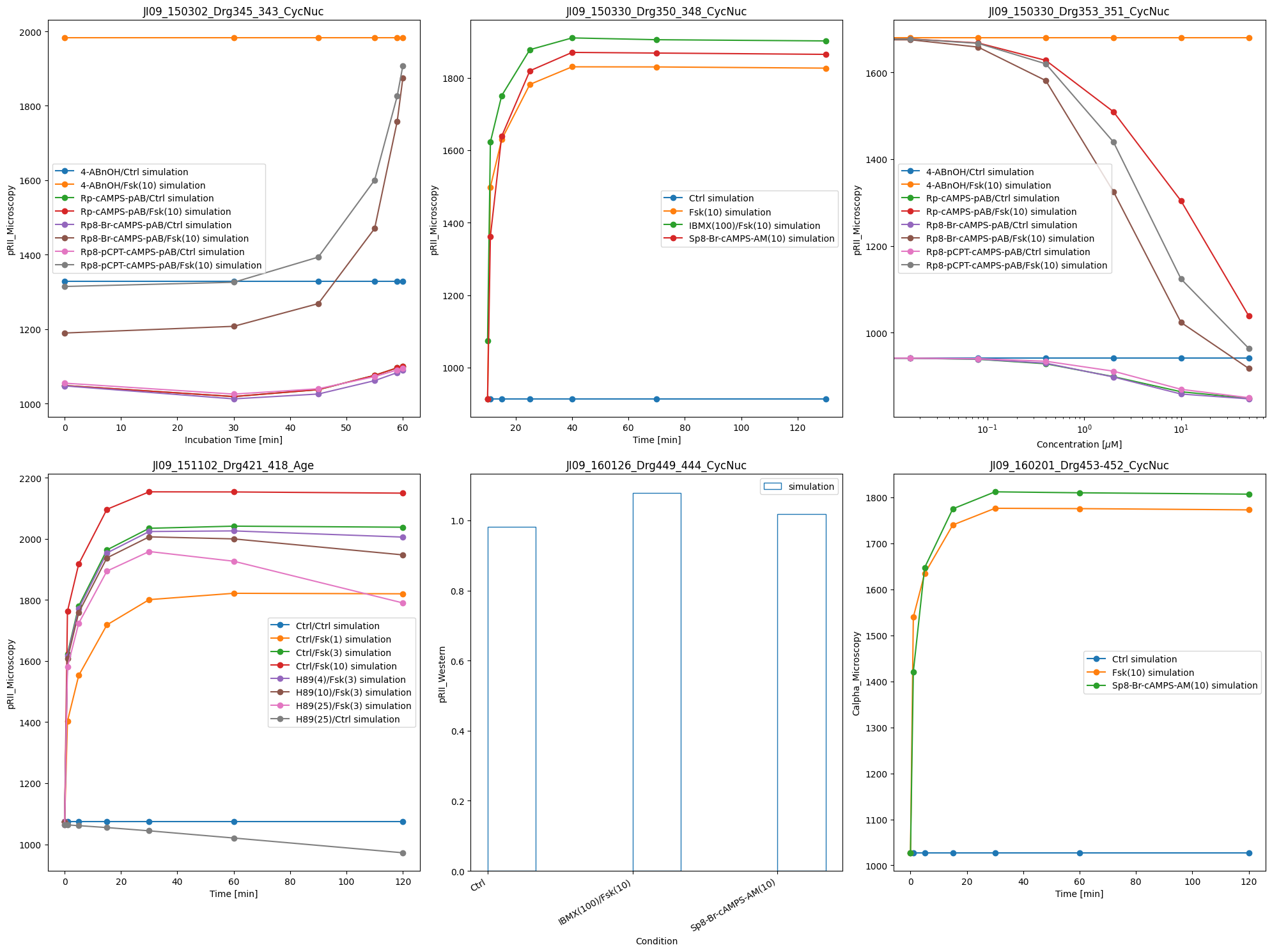
One can also plot only simulated data:
ax = plot_with_vis_spec(
visualization_file_path,
condition_file_path,
simulations_df=simulation_file_path,
)
If both measurements and simulated data are available, they can be visualized using scatter plot:
visualization_file_scatterplots = (
folder + "Isensee_visualizationSpecification_scatterplot.tsv"
)
ax = plot_with_vis_spec(
visualization_file_scatterplots,
condition_file_path,
data_file_path,
simulation_file_path,
)
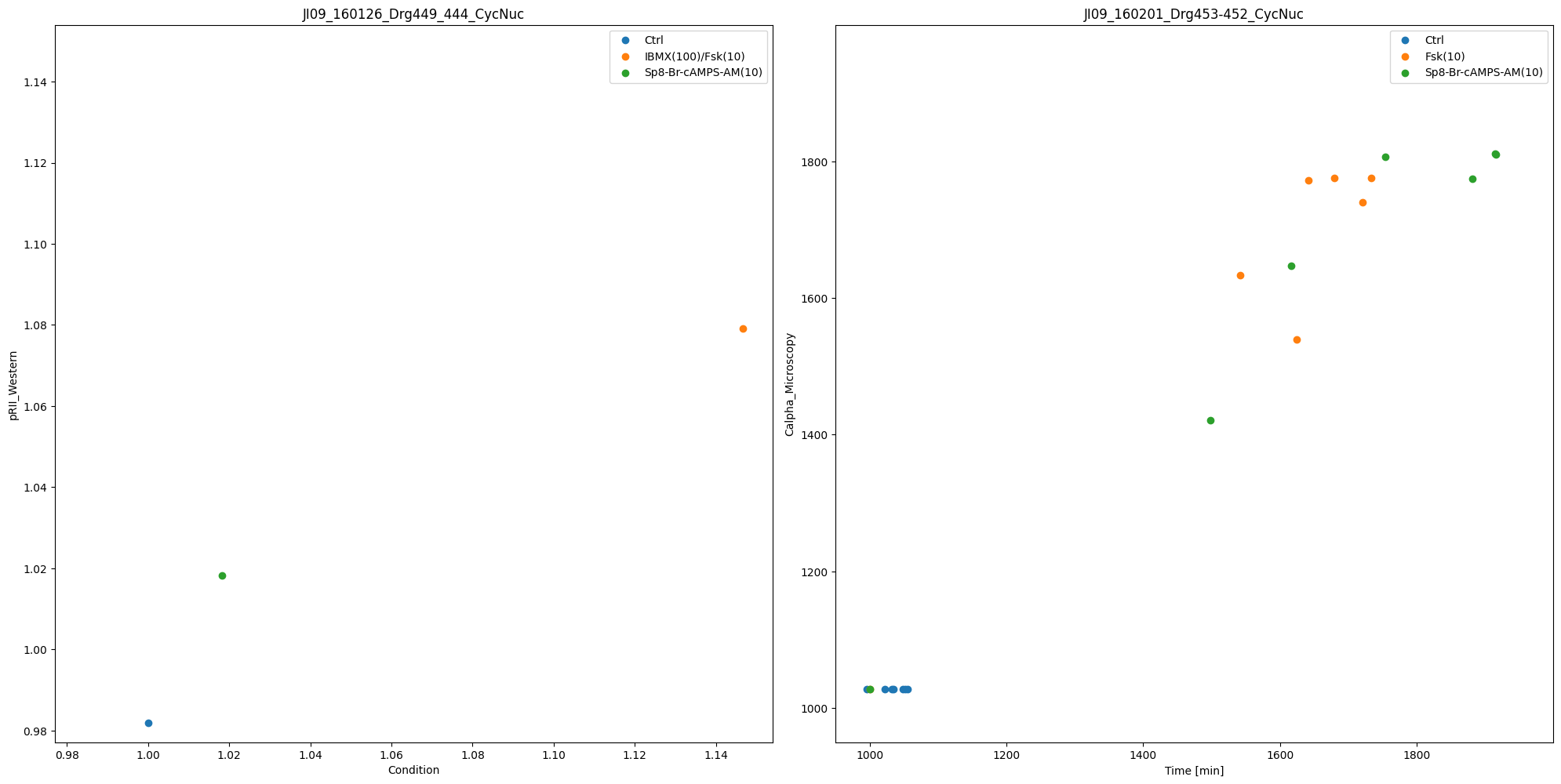
We can also call the plotting routine without the visualization specification file, but by passing a list of lists as dataset_id_list. Each sublist corresponds to a plot, and contains the datasetIds which should be plotted.
In this simply structured plotting routine, the independent variable will always be time.
datasets = [
[
"JI09_150302_Drg345_343_CycNuc__4_ABnOH_and_ctrl",
"JI09_150302_Drg345_343_CycNuc__4_ABnOH_and_Fsk",
],
[
"JI09_160201_Drg453-452_CycNuc__ctrl",
"JI09_160201_Drg453-452_CycNuc__Fsk",
"JI09_160201_Drg453-452_CycNuc__Sp8_Br_cAMPS_AM",
],
]
ax_without_sim = plot_without_vis_spec(
condition_file_path, datasets, "dataset", data_file_path
)
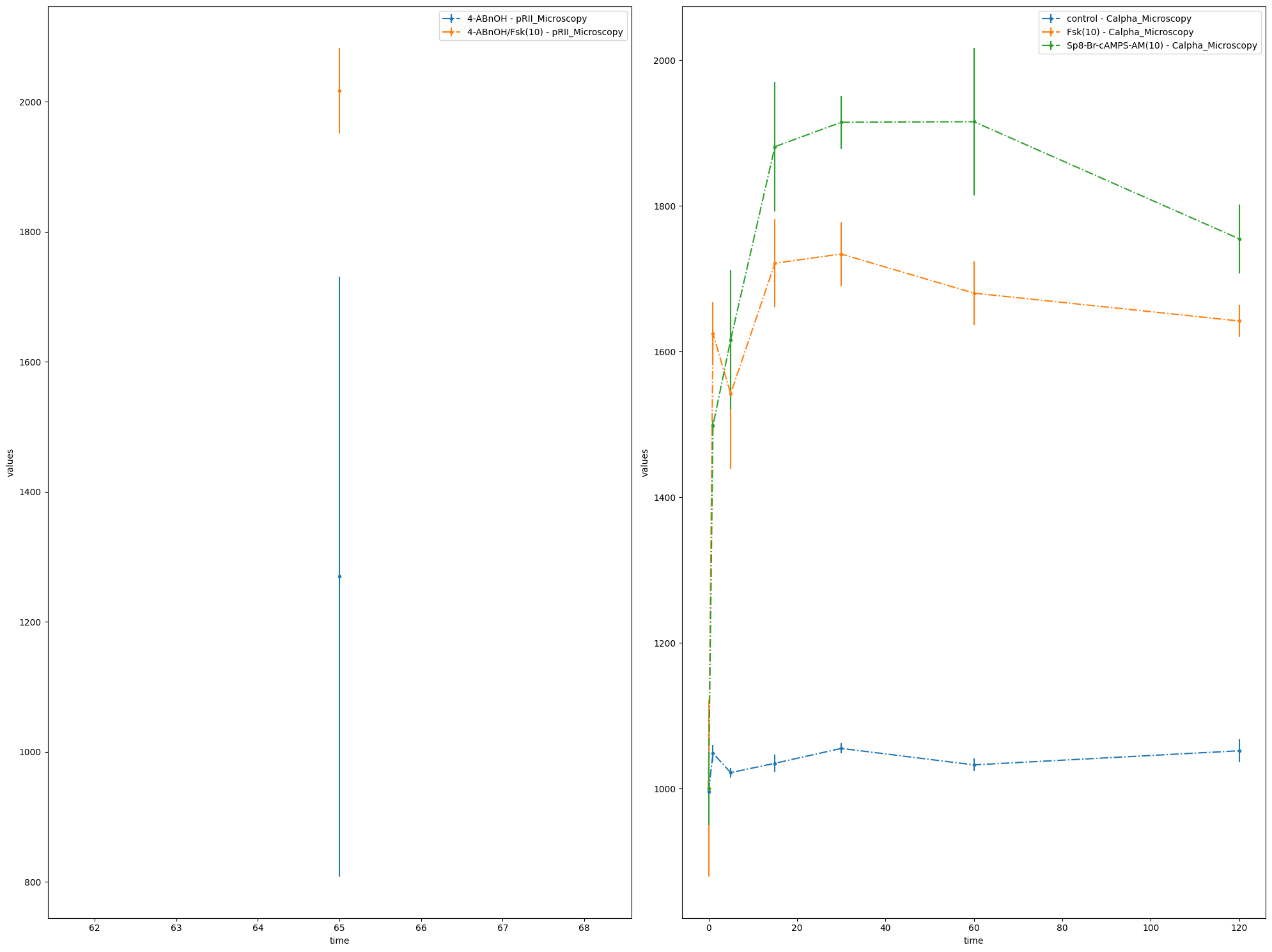
Let’s look more closely at the plotting routines, if no visualization specification file is provided. If such a file is missing, PEtab needs to know how to group the data points. For this, three options can be used:
dataset_id_list
sim_cond_id_lis
observable_id_list
Each of them is a list of lists. Again, each sublist is a plot and its content are either simulation condition IDs or observable IDs or the dataset IDs.
We want to illustrate this functionality by using a simpler example, a model published in 2010 by Fujita et al.
data_file_Fujita = "example_Fujita/Fujita_measurementData.tsv"
condition_file_Fujita = "example_Fujita/Fujita_experimentalCondition.tsv"
# Plot 4 axes objects, plotting
# - in the first window all observables of the simulation condition 'model1_data1'
# - in the second window all observables of the simulation conditions 'model1_data2', 'model1_data3'
# - in the third window all observables of the simulation conditions 'model1_data4', 'model1_data5'
# - in the fourth window all observables of the simulation condition 'model1_data6'
sim_cond_id_list = [
["model1_data1"],
["model1_data2", "model1_data3"],
["model1_data4", "model1_data5"],
["model1_data6"],
]
ax = plot_without_vis_spec(
condition_file_Fujita,
sim_cond_id_list,
"simulation",
data_file_Fujita,
plotted_noise="provided",
)
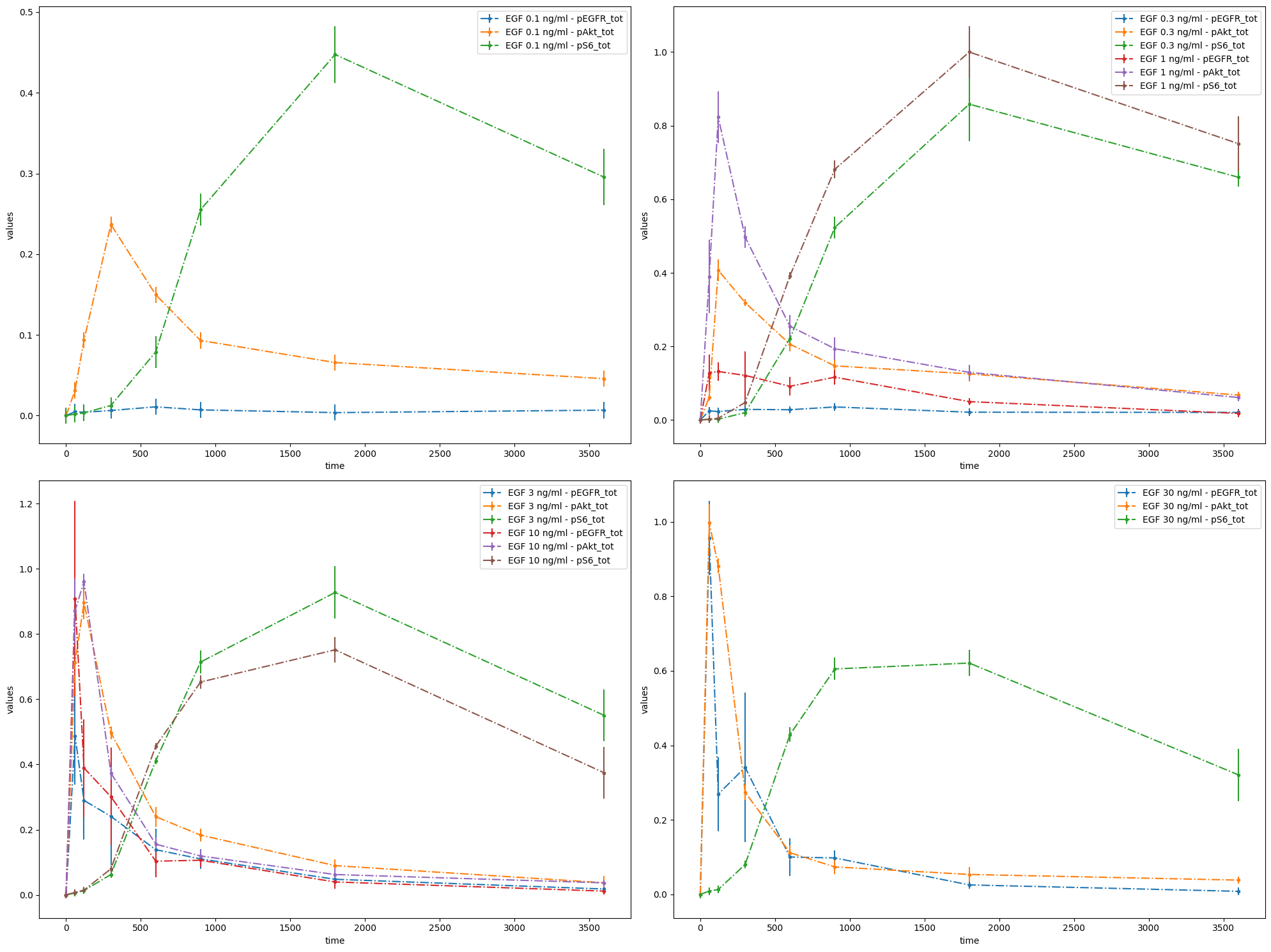
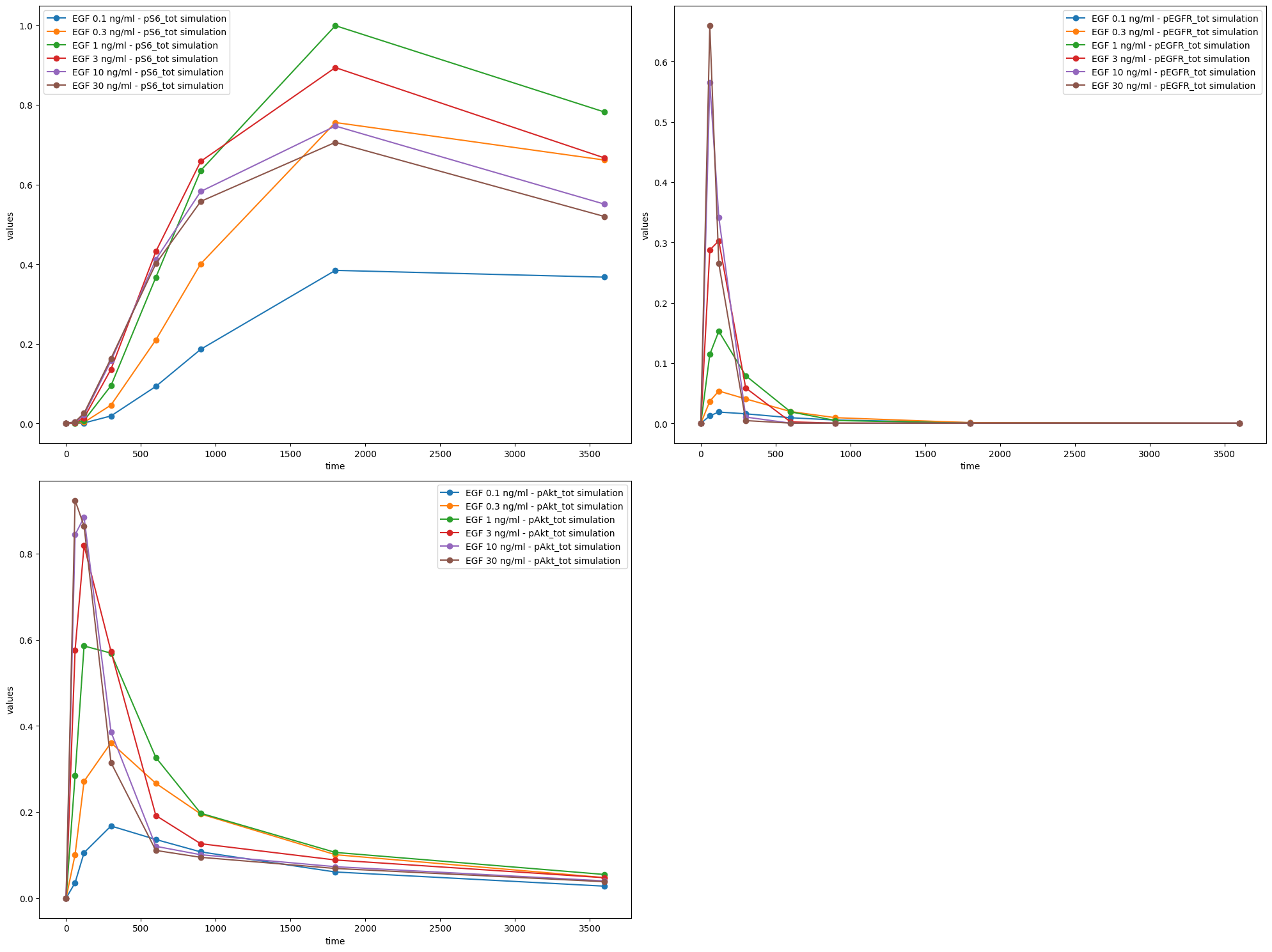
# Plot 3 axes objects, plotting
# - in the first window the observable 'pS6_tot' for all simulation conditions
# - in the second window the observable 'pEGFR_tot' for all simulation conditions
# - in the third window the observable 'pAkt_tot' for all simulation conditions
observable_id_list = [["pS6_tot"], ["pEGFR_tot"], ["pAkt_tot"]]
ax = plot_without_vis_spec(
condition_file_Fujita,
observable_id_list,
"observable",
data_file_Fujita,
plotted_noise="provided",
)
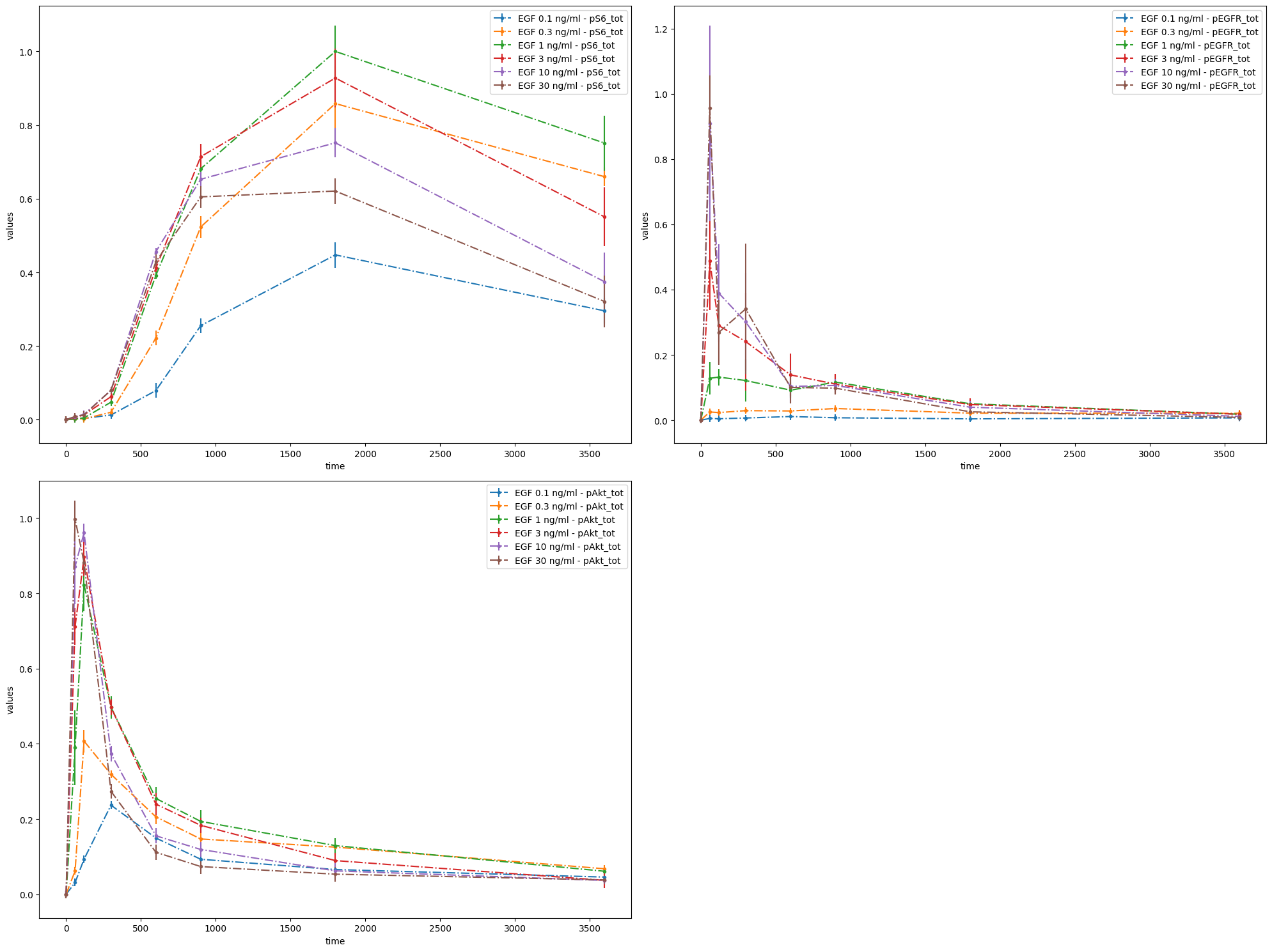
# Plot 2 axes objects, plotting
# - in the first window the observable 'pS6_tot' for all simulation conditions
# - in the second window the observable 'pEGFR_tot' for all simulation conditions
# - in the third window the observable 'pAkt_tot' for all simulation conditions
# while using the noise values which are saved in the PEtab files
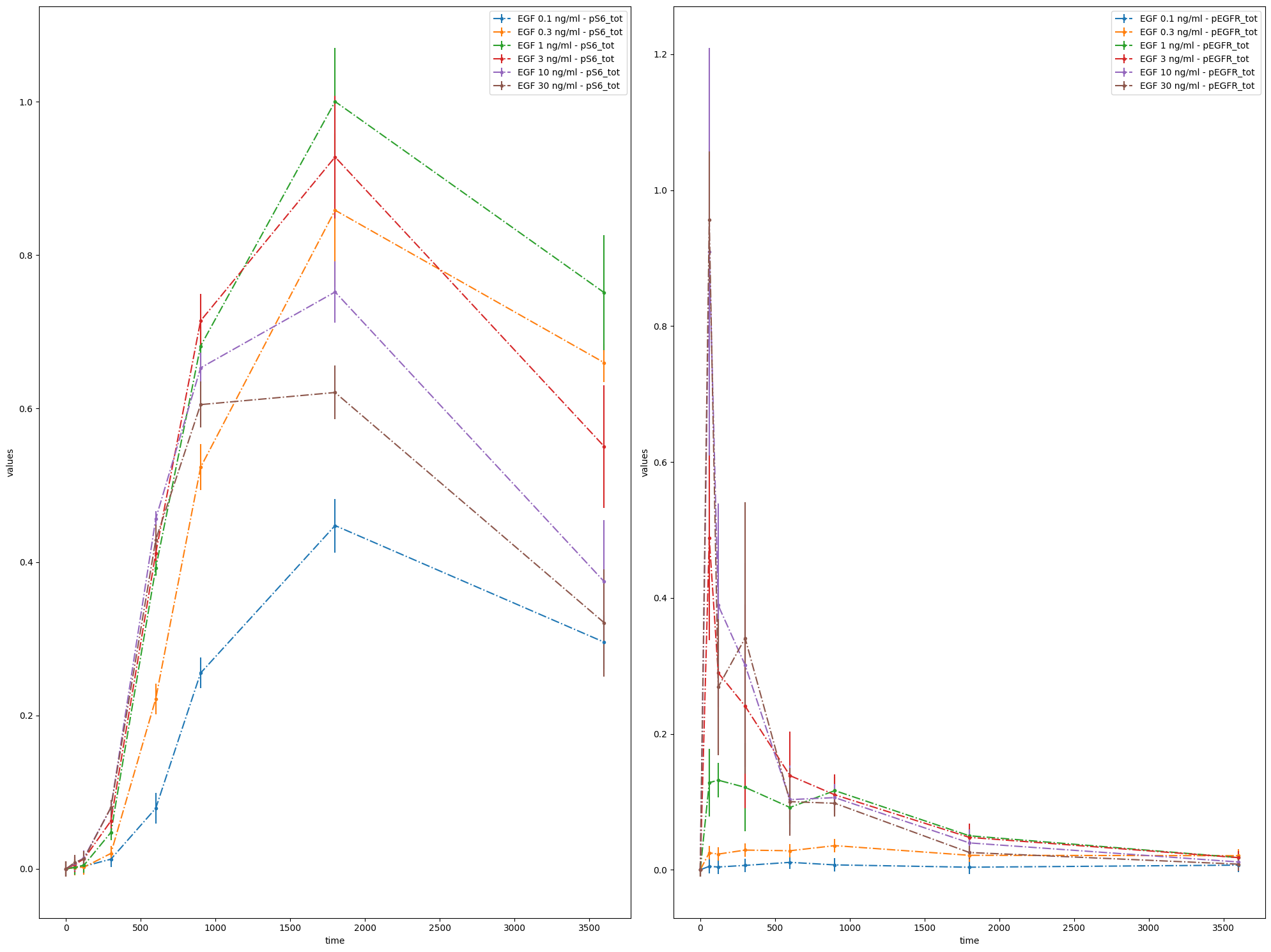
observable_id_list = [["pS6_tot"], ["pEGFR_tot"]]
ax = plot_without_vis_spec(
condition_file_Fujita,
observable_id_list,
"observable",
data_file_Fujita,
plotted_noise="provided",
)
Plot only simulations
simu_file_Fujita = "example_Fujita/Fujita_simulatedData.tsv"
sim_cond_id_list = [
["model1_data1"],
["model1_data2", "model1_data3"],
["model1_data4", "model1_data5"],
["model1_data6"],
]
ax = plot_without_vis_spec(
condition_file_Fujita,
sim_cond_id_list,
"simulation",
simulations_df=simu_file_Fujita,
plotted_noise="provided",
)
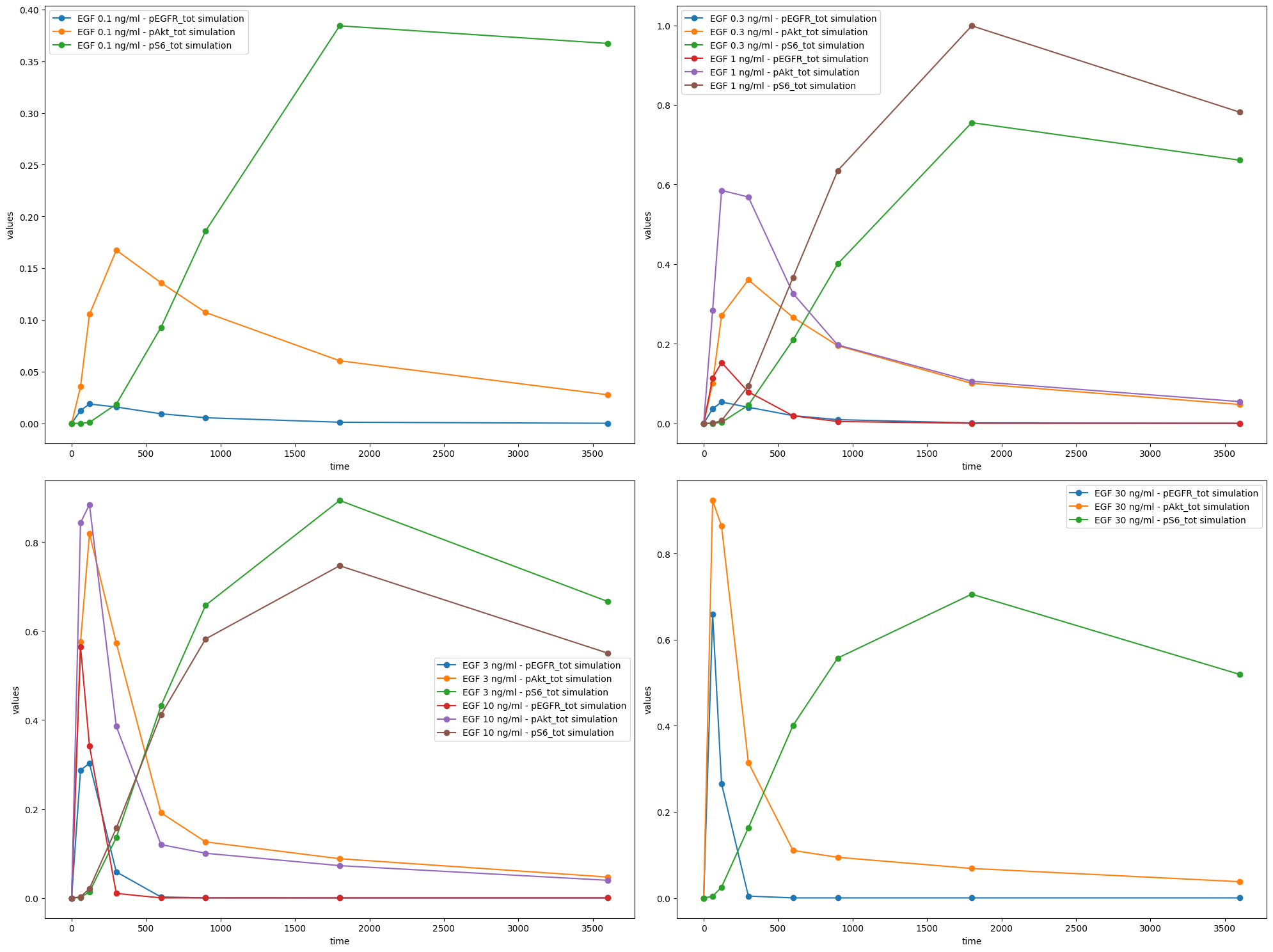
observable_id_list = [["pS6_tot"], ["pEGFR_tot"], ["pAkt_tot"]]
ax = plot_without_vis_spec(
condition_file_Fujita,
observable_id_list,
"observable",
simulations_df=simu_file_Fujita,
plotted_noise="provided",
)